David Brow develops improved method for tracking molecular signals in living cells
A University of Wisconsin-Madison researcher has established a new approach to understanding how protein and RNA molecules in a cell cooperate to direct the proper expression of genes.
The approach improves upon a well-established method called genetic suppression, developed more than a century ago. A genetic suppressor is a second mutation that corrects the defect of a primary mutation, according to David Brow, PhD, professor of biomolecular chemistry.
The research was recently published in the journal Genetics.
By using a customized targeting sequencing technique, Brow found that he could identify suppressor mutations much more easily and in much larger numbers than before.
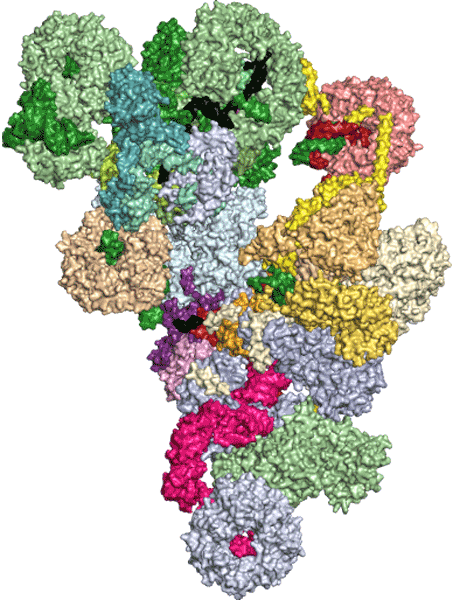
By mapping the mutations on existing molecular structures, he obtained new insights into an essential step of gene expression called pre-messenger RNA splicing.
“This approach should be broadly useful for understanding the dynamics of many different complex biological assemblies or pathways,” Brow said.
The new research targeted a specific cold-sensitive mutation, U4-cs1, that blocks activation of the spliceosome, essentially an incredibly small machine within the nucleus of a cell. It is made of RNA and protein and removes meaningless information from cellular RNAs.
The approach identified five new genes in which mutations can overcome the block created by U4-cs1.
RNA splicing is one of the fundamental processes in gene expression for cells in which DNA exists inside a separate compartment called the nucleus. Organisms that have a nucleus are called eukaryotes. Brow’s research used brewer’s yeast, but humans also are eukaryotes and have a very similar spliceosome.
Proper function of the spliceosome is essential for all eukaryotic cells. Many human disease mutations prevent proper splicing of a pre-messenger RNA by the spliceosome.
To identify the suppressor mutations, Brow’s system relied upon a custom sequencing panel commissioned from Illumina, Inc., that targeted 112 genes encoding pre-messenger RNA splicing factors in the genome of yeast. Assembly of all of these factors into a functioning spliceosome requires a dynamic series of interactions between the components called an “allosteric cascade.”
By identifying and better understanding suppressors of the U4-cs1 mutation, Brow hopes to learn how the components of the spliceosome signal to each other to coordinate the steps of pre-mRNA splicing, he said. This understanding may allow scientists to correct the splicing defects caused by inherited mutations that lead to diseases.
“This approach shows great promise for understanding the myriad of molecular interactions that are required for accurate and efficient pre-mRNA splicing by the spliceosome,” Brow said.